Running simulations on HPC
At the end of this class, you should be able to:
- Organize and plan simulation files on the cluster
- Run the simulation and optimize
- Perform a run-time analysis of flow parameters
Now we are ready to start a large-scale CFD simulation. We have generated the mesh, setup the simulation, and run scaling tests. The purpose of this section is to standardize the workflow of organizing, running, and monitoring large-scale CFD simulations on a remote HPC system and to provide best-practice tips.
- All examples will be carried out on Graham, but the approach is generalizable to any HPC system
- The workflow is NOT set in stone, but rather suggestions to facilitate HPC usage
- The examples will be performed in both OpenFOAM and SU2; Students can toggle between OpenFOAM and SU2 as shown below.
OpenFOAM commands
SU2 commands
Remote files on the cluster
Before running a simulation, it is good practice to organize the file system on the remote cluster. Although this step may be time-consuming to implement, it will save time in the long run. The question now is: how do we organize our simulation files?
To answer this question, let us first figure out what options we have. Upon logging into Graham, type:
[user@gra-login1] diskusage_reportThis command will check the available disk space and the current disk utilization on our personal and group profiles. The output will look something like:
[user@gra-login1] Description Space # of files /home (user username) 23G/50G 112/500k /scratch (user username) 6633G/20T 14k/1000k /project (group username) 0/2048k 0/1025 /project (group def-piname) 292M/1000G 294/505k /project (group rrg-piname-ac) 49T/100T 112k/505kWhere username refers to your personal space, while piname refers to your group (or principal investigator) profile.
/homehas a small capacity which is suitable for code development, source code, small parameter files, job submission scripts and version control. Note that we cannot write to the/homedrive from the compute nodes./project (group rrg-piname-ac)is a directory that is linked to your principal investigator’s account and meant for longer-term storage and sharing data among members of a research group./scratchis connected to a single user, and is intended for intensive read/write operations on large files. As mentioned in section 2.1, it is the right place to set up and run your simulations.
More detailed information on the Alliance’s storage and management systems can be found on their website.
Important files must be copied off /scratch regularly since they are not backed up and older files are subject to purging!
As we have space on the /home drive, if it’s not already done, we can clone from the course GitHub repository. The repository contains the input files and mesh for the examples (and is of modest size), therefore the /home drive will allow us to modify and save the examples prior to copying the cases to /scratch for running the simulations.
[user@gra-login1] lsREADME case movie.ogv run.shbfs_Umag.pvsm mesh run_jobscript.sh[user@gra-login1] lsCoarse READMEFine ScalingIntermediate Turbulent-flow-over-Backward-facing-step.pdfCreate the run directory
Now that you have cloned the repository to your /home directory, we can make any modifications. Any modifications you want to do on the source should happen in /home, as it’s your own personal space, and nothing will be purged from here.
For this reason, once you are satisfied with the changes implemented in the source code, you should copy the code into a run directory in /scratch. In this case, no changes were required in the source files, therefore we can copy them directly:
[user@gra-login1] cp -r * ./scratch/01_BFS_openFOAM[user@gra-login1] cp -r ./Coarse/ ./scratch/02_BFS_SU2Naming convention and folder structure
At this stage, do not underestimate the importance of naming conventions for files and directories. Consistent naming conventions can help you find your data, avoid mistakes, and minimize duplication of efforts. Some CFD codes, such as OpenFoam, have strict folder naming and structure conventions for each simulation, while other codes, such as SU2, do not. As each CFD project typically comprises multiple simulations (mesh refinement, scaling tests, different turbulence models), a consistent naming convention and folder structure, determined a priori, will facilitate the organization, running, postprocessing, and, ultimately, the research data management.
Best practices in folder naming conventions:
- Avoid space and special characters in the names!: Use either dashes (-) or underscores (_) if you need to separate elements in the folder name. Alternatively, you can use the camelCase in which the first word is in lowercase and every other word starts with a capital letter (e.g. pimpleFoam).
- Keep it short, but meaningful: Keep folder names as short as possible and consider using abbreviations (write those down!)
- Write down the naming convention: Write down the naming convention in the data management plan (DMP), more in Section 3, or in a README file.
- Dates in folder names should follow an ISO 8601 format If dates are used in the folder names, use a well-accepted standard format YYYYMMDD or YYYY-MM-DD.
Here are a couple considerations for directory naming conventions:
-
Think about the simulations you plan to run: Consider all the possible simulations that you will need to run:
- What are the important parameters of the simulations? Here are some typical parameters that may be varied between simulations:
- turbulence models (SST, k-omega, RSM etc.)
- boundary conditions (freestream velocity, wall resolved, wall modelled, Reynolds number)
- various grid resolutions (coarse, medium, fine, etc.)
- thermophysical properties (Prandtl number, thermal convection, etc.)
- …
- What are the important parameters of the simulations? Here are some typical parameters that may be varied between simulations:
-
Establish a consistent naming convention and folder structure
- Depending on the planned parameter space that you plan to cover, you can establish a consistent naming convention and folder structure. Depending on the complexity of the CFD project, you may opt for either:
- a more complex folder hierarchy
- a more comprehensive folder naming convention
The naming convention and folder structure need not be unique and different research projects may have different naming convention. Here are two different examples:
- Depending on the planned parameter space that you plan to cover, you can establish a consistent naming convention and folder structure. Depending on the complexity of the CFD project, you may opt for either:
Having a deeper folder and subfolder hierarchy may help to organize the various simulations, especially for larger CFD projects. As the simulation organization is embedded within the folder structure, the naming convention of each folder is less critical. Here is an example:
./BackwardFacingStep|-- gridConvergenceStudy/| +-- coarse/| +-- medium/| +-- fine/| +-- README|-- workingDir/|-- scalingTests/| +-- weakScaling/| | +-- ...| +-- strongScaling/| | +-- Sim_40proc/| | +-- Sim_20proc/| | +-- ...|-- turbulenceModels/| +-- SST/| +-- kOmega/| +-- SST/|-- READMEFor smaller CFD projects, a flatter folder structure may be preferred to facilitate navigation among various simulations. The flatter folder structures come at the cost of a very comprehensive naming convention. Here is an example:
[SIMULATION_NAME]_[Reynolds_number]_[Turbulence_model]_[resolution]
where [SIMULATION_NAME] is BFS for the backward facing step, [Reynolds_number] is ReXXXX where XXXX is the Reynolds number, [Turbulence_model] defines the turbulence models (SST, KOM: k-omega, RSM: Reynolds stress modelling etc.), and [resolution] is the resolution of the mesh (CRS coarse, MDM medium, FIN fine etc.). Based on these conventions
BFS_Re5000_SST_CRS
BFS_Re5000_SST_CRS
./BackwardFacingStep|-- BFS_Re5000_SST_CRS/|-- BFS_Re5000_SST_MDM/|-- BFS_Re5000_SST_FIN/|-- workingDir/|-- BFS_Re5000_KOM_FIN/|-- BFS_Re5000_RSM_FIN/|-- BFS_Re1000_KOM_FIN/|-- BFS_Re10000_KOM_FIN/|-- READMEBookkeeping
When running a large number of simulations, in addition to a consistent naming convention and folder structure, it is good practice to maintain a centralized database of the simulations. This database, which could be an Excel sheet, can provide a quick summary of the important details (date of simulation, number of processors, code compilation characteristics, etc.) for each simulation and any user comment.
Setting up simulation parameters
As the tutorial files have already been prepared, here we highlight only the most important steps in setting up the files prior to running the simulations. We assume that we are using a mesh with a known resolution that has been generated by an external meshing tool (see details in section 2.4). For simplicity and ease of computation, we only use the coarsest mesh with about bfs_200k_DDES in OpenFoam, and Coarse in SU2 examples). With this mesh, we can set the remaining parameters of the simulations:
Simulation setups can be daunting. Fortunately, we rarely need to construct a simulation setup from scratch. Instead, the best practice is to first test cases which are often provided within the CFD package. For example, we can look at:
You can then select the closest case to the one you plan to simulate and adapt it. Selecting the best case among those available is often not trivial.
-
Initial and boundary conditions: TODO
-
Numerics: the selection of the numerical details for both the temporal and spatial calculations will directly impact the computational cost. The specific details on the selection of the numerics falls outside the scope of ARC4CFD, therefore it is only a list of some of the considerations to set:
- Explicit, Semi-implicit, and Implicit time advancement and/or the order of the selected numerical scheme
- Spatial discretization and order of scheme (convective and diffusive terms, we can also set details for turbulence equations)
- Pre-conditioning scheme (Jacobi, ILU, LU_SGS etc.)
- Type of linear solvers (FGMRES, BCSTAB, etc.)
- Stabilization schemes
- Convergence criteria
- …
-
Time step size: The chosen numerics, the local grid resolution, and the minimal resolved time scale will directly impact the time step of the simulation. In the present tutorial case, with an explicit time advancement, the time step is CFL bound (section 2.3) and requires:
. Alternatively, in most CFD codes, we can set the maximum CFL number, and the solver will select the to meet the CFL conditions. The advantage of fixing is that we know that we evolved to 1 s after 10,000 time steps, whereas fixing the CFL allows us to maximize the based on the stability of the code.
OpenFOAM
vim case/system/controlDict# change deltaT to 1e-4;SU2
vim Coarse/Backstep_str_config.cfg# change TIME_STEP (line 118)- Simulation End Time: For steady CFD simulations, for a given set of boundary conditions, the residual of the simulation must be reduced to the desired convergence criterion. For unsteady CFD simulations, especially with turbulence and/or geometric complexities, it is important to run the simulation long enough to let the flow properly develop in the computational domain. As the flow adapts from it’s initial conditions given the boundary conditions of the problem, there will inevitably be a transient phase. Eventually, the flow will reach a statistically steady state during which the spatially- or phased-averaged statistical quantities (drag coefficient, turbulent fluctuation etc.) remain constant. As mentioned in section 2.3 estimating the time required to reach a steady state is very difficult and is case dependent. A reasonably good measure to get a rough estimate is the flow through time (FTT) as described in section 2.2. In this example, we choose the end time to correspond to
flow-through times.
OpenFOAM
vim case/system/controlDict# change endTime to 0.5;SU2
vim Coarse/Backstep_str_config.cfg# change TIME_ITER (line 120) to 5000- Output and snapshot time interval: for the flow analysis, we typically rely on a combination of:
- run-time statistics: these statistics are collected during run time
- postprocessed statistics: these statistics are postprocessed after the simulation from the output data
Although run time statistics are often desired as we can get high temporal resolution, they can impart a significant run time penalty on large simulations (e.g. averaging on a plane). Postprocessed statistics provide more flexibility (we can compute new statistics even after the simulation has been run.) but demand significantly more flow realizations or snapshots compute.
OpenFOAM
vim case/system/controlDict# change writeInterval to 20;SU2
vim Coarse/Backstep_str_config.cfg# change OUTPUT_WRT_FREQ to OUTPUT_WRT_FREQ = 4800, 20# this will make sure that restart files are written before# the simulation end time is reached, and snapshots are saved every 20 steps.- Domain decomposition: after performing the scaling test for a given mesh (section 2.5), we know how many processors we should use to optimize the CFD workflow.
OpenFOAM
The user should modify the numberOfSubdomains entry in the case/system/decomposeParDict file. In this example, we use 64 processors.
vim case/system/decomposeParDict# change numberOfSubdomains to 64;(depending of on the selected parallelization method (see openFoam user guide) there may be other parameters to be modified)
SU2
No need to specify this a priori in SU2. During execution, the user will decide how many processors to use for the calculation.
After all flow and simulation parameters have been set, we are now ready to run the simulation.
Run a large-scale CFD simulation
As previously seen in section 1.5, when solving the two-dimensional Poisson equation, there are 2 common ways of running large-scale simulations on the cluster:
- an interactive session by logging into the compute nodes directly, and
- submitting a batch job to SLURM. Interactive sessions are easier to set up and debug, as we can interactively run the simulation on the compute node and immediately assess the outputs. But the interactive sessions are not suited for long jobs (as the terminal window must remain open and the workstation on), many processors, or multiple parallel simulations. Therefore, interactive sessions should only be used for small simulations, debugging large simulations, and/or running scaling tests.
Now that we copied the tutorial case from our /home directory onto /scratch, our goal is to run 3 simulations starting from a coarse mesh (
Running in interactive mode
- Allocate required HPC resources:
salloc -n 64 --time=10:00:0 --mem-per-cpu=3g --account=account-name- Create the sub-case directory
bfs_200k_DDESwithin the main case directory:
[user@gra796] cp -r * ./case/* ./bfs_200k_DDES- Generate mesh from file using the
gmshutility:
[user@gra796] gmsh mesh/bfs_200k.geo -3 -o mesh/bfs_200k.msh -format msh2- Convert mesh to OpenFOAM format and modify boundary file to reflect boundary conditions:
[user@gra796] gmshToFoam mesh/bfs_200k.msh -case /bfs_200k_DDES[user@gra796] cp /bfs_200k_DDES/constant/polyMesh/boundary /bfs_200k_DDES/constant/polyMesh/boundary.old[user@gra796] sed -i '/physical/d' /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i "/wall_/,/startFace/{s/patch/wall/}" /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i "/top/,/startFace/{s/patch/symmetryPlane/}" /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i "/front/,/startFace/{s/patch/cyclic/}" /bfs_200k_DDES/constant/polyMesh boundary[user@gra796] sed -i "/back/,/startFace/{s/patch/cyclic/}" /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i -e '/front/,/}/{/startFace .*/a'"\\\tneighbourPatch back;" -e '}' /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i -e '/back/,/}/{/startFace .*/a'"\\\tneighbourPatch front;" -e '}' /bfs_200k_DDES/constant/polyMesh/boundary[user@gra796] sed -i -e '/cyclic/,/nFaces/{/type .*/a'"\\\tinGroups 1(cyclic);" -e '}' /bfs_200k_DDES/constant/polyMesh/boundary[user@ggra796] sed -i -e '/wall_/,/}/{/type .*/a'"\\\tinGroups 1(wall);" -e '}' /bfs_200k_DDES/constant/polyMesh/boundary- Start the simulation:
[user@gra796] cd ./bfs_200k_DDES[user@gra796] ./AllrunWhere the Allrun script performs some very important operations. Among them:
cp -r 0.orig 0 # initialize the flowrunApplication decomposePar # decomposed the domain based on the # of procs.runParallel $(getApplication) # run the application in parallelrunApplication reconstructPar # after simulation is done join processors into a single filerm -rf processor* # remove all the single processors directoriesAfter step 5 is completed, you should see the simulation starting on the terminal:
Running decomposePar on /home/ambrox/scratch/BFS_OpenFOAM/bfs_200k_DDESRunning pimpleFoam in parallel on /home/ambrox/scratch/BFS_OpenFOAM/bfs_200k_DDES using 64 processesAt this point, the terminal window hangs while the simulation runs. If the terminal window is closed, the simulation stops.
- Allocate required HPC resources:
salloc -n 64 --time=10:00:0 --mem-per-cpu=3g --account=account-name- Generate mesh from file using the
gmshutility:
[user@gra796] gmsh Backstep_str_mesh.geo -0- Start the simulation:
[user@gra796] mpirun -n 64 SU2_CFD Backstep_str_config.cfgSubmitting a batch script
When dealing with multiple simulations, long duration, or a large number of processors, it is best to submit SLURM. As seen earlier, SLURM will queue the job and run it when the resources become available. In this case, for instance, we could include Steps 1-5 in a single file run.sh to be run in interactive mode, or even better in a batch job script run_jobscript.sh to submit to the job scheduler. Both files are included in the GitHub repository and are shown below:
run.sh
#uncomment only one of the following three lines to select the meshmesh=bfs_200k # coarse mesh#mesh=bfs_400k # intermediate mesh#mesh=bfs_800k # fine mesh
case="$mesh"_DDESif [ ! -d "$case" ]; then echo "creating $case"fi
cp -r case $casegmsh mesh/$mesh.geo -3 -o mesh/$mesh.msh -format msh2gmshToFoam mesh/$mesh.msh -case $casecp $case/constant/polyMesh/boundary $case/constant/polyMesh/boundary.old
#### script to modify boundary file to reflect boundary conditions############# sed -i '/physical/d' $case/constant/polyMesh/boundary sed -i "/wall_/,/startFace/{s/patch/wall/}" $case/constant/polyMesh/boundary sed -i "/top/,/startFace/{s/patch/symmetryPlane/}" $case/constant/polyMesh/boundary sed -i "/front/,/startFace/{s/patch/cyclic/}" $case/constant/polyMesh/boundary sed -i "/back/,/startFace/{s/patch/cyclic/}" $case/constant/polyMesh/boundary sed -i -e '/front/,/}/{/startFace .*/a'"\\\tneighbourPatch back;" -e '}' $case/constant/polyMesh/boundary sed -i -e '/back/,/}/{/startFace .*/a'"\\\tneighbourPatch front;" -e '}' $case/constant/polyMesh/boundary sed -i -e '/cyclic/,/nFaces/{/type .*/a'"\\\tinGroups 1(cyclic);" -e '}' $case/constant/polyMesh/boundary sed -i -e '/wall_/,/}/{/type .*/a'"\\\tinGroups 1(wall);" -e '}' $case/constant/polyMesh/boundary#### script to modify boundary file to reflect boundary conditions#############
#cp mesh/boundary $case/constant/polyMesh/cd $caserm -r log.* proc*./Allrunrun_jobscript.sh
#!/bin/bash#SBATCH --job-name="bfs_DDES" # job name
#SBATCH --ntasks=64 # number of processors#SBATCH --nodes=2-8 # number of nodes#SBATCH --mem-per-cpu=3g # memory per cpu
#SBATCH --time=0-20:00 # walltime dd-hh-mm#SBATCH --output=bfs%j.txt # output file#SBATCH --mail-type=FAIL
cd $SLURM_SUBMIT_DIRmodule load StdEnv/2023 gcc/12.3 openmpi/4.1.5 openfoam/v2306 gmsh/4.12.2. $WM_PROJECT_DIR/bin/tools/RunFunctions
#Before running this script#---------------------------------------------------------------------------------------------------------------# 1) Uncomment only one of the three lines under MESH to select the mesh;# 2) Replace 64 against "numberOfSubdomains: in 'system/decomposeParDict' of base_case with no.of tasks (= nodes * tasks per node)# 3) Use following deltaT in 'system/controlDict' in your base_case (You might want to change the writing interval too)#---------------------------------------------------------------------------------------------------------------
#------------ USER INPUT START ------------------------------------------------------
# BASE CASEbase_case=case # Provide name of the base case with your preferred settings. # prepare your base case from the supplied "case" folder
# MESHmesh=bfs_200k # coarse mesh (deltaT: 1e-4)#mesh=bfs_400k # intermediate mesh (deltaT: 4e-5)#mesh=bfs_800k # fine mesh (deltaT: 2e-5)
#------------ USER INPUT END ------------------------------------------------------
case="$mesh"_DDESif [ ! -d "$case" ]; then # This checks if a directory exists with the same case name. Helps for restarting a job echo "creating $case" cp -r $base_case $case #copying from base case gmsh mesh/$mesh.geo -3 -o mesh/$mesh.msh -format msh2 #generating mesh gmshToFoam mesh/$mesh.msh -case $case #converting mesh to openfoam format
#### script to modify constant/polyMesh/boundary file to reflect boundary conditions############# sed -i '/physical/d' $case/constant/polyMesh/boundary sed -i "/wall_/,/startFace/{s/patch/wall/}" $case/constant/polyMesh/boundary sed -i "/top/,/startFace/{s/patch/symmetryPlane/}" $case/constant/polyMesh/boundary sed -i "/front/,/startFace/{s/patch/cyclic/}" $case/constant/polyMesh/boundary sed -i "/back/,/startFace/{s/patch/cyclic/}" $case/constant/polyMesh/boundary sed -i -e '/front/,/}/{/startFace .*/a'"\\\tneighbourPatch back;" -e '}' $case/constant/polyMesh/boundary sed -i -e '/back/,/}/{/startFace .*/a'"\\\tneighbourPatch front;" -e '}' $case/constant/polyMesh/boundary sed -i -e '/cyclic/,/nFaces/{/type .*/a'"\\\tinGroups 1(cyclic);" -e '}' $case/constant/polyMesh/boundary sed -i -e '/wall_/,/}/{/type .*/a'"\\\tinGroups 1(wall);" -e '}' $case/constant/polyMesh/boundary #### script to modify constant/polyMesh/boundary file to reflect boundary conditions#############
cd $case rm -r log.* proc* cp -r 0.orig 0
decomposePar -force > log.decomposePar cd ..fi
cd $casempirun pimpleFoam -parallel > log.pimpleFoamreconstructPar > log.reconstructPar#rm -rf processor*su2job_StdEnv.sh
#!/bin/bash#SBATCH --nodes=1#SBATCH --ntasks-per-node=10#SBATCH --time=0-23:00#SBATCH --job-name ARC4CFD_BFS#SBATCH --output=BFSstr%j.txt#SBATCH --mail-type=FAILcd $SLURM_SUBMIT_DIRmodule load StdEnv/2020 gcc/9.3.0 openmpi/4.0.3 su2/7.5.1mpirun -n 10 SU2_CFD Backstep_str_config.cfgThe command to submit the batch script is simply:
[user@gra-login1] sbatch run_jobscript.shSubmitted batch job 26236582[user@gra-login1] sbatch su2job_StdEnv.shSubmitted batch job 26236582QUIZ
Run the numerical simulation of the same backward facing step flow for the mesh containing
2.6.1 What would be the time step size
2.6.2 What would be the writeInterval required to still print results every 2 milliseconds?
Perform a runtime analysis of the simulation
Once the job is submitted, we should make sure the simulation is running properly. This is done by typing the command:
[user@gra-login1] sq
JOBID USER ACCOUNT NAME ST TIME_LEFT NODES CPUS TRES_PER_N MIN_MEM NODELIST (REASON)26236582 username def-piname bfs_DDES R 19:59:46 8 64 N/A 3G gra[11005,11007,11010,11012-11016] (None)Based on the output, the code is running, as expected, on 64 processors using 8 nodes. This check, however, does not really tell us that everything is going well, but only that the 64 processes have started and are working on something. The next step would be to check the log file.
If you recall, with the command mpirun pimpleFoam -parallel > log.pimpleFoam in the run_jobscript.sh we asked the code to write any output to a log file called log.pimpleFoam. If you notice, this file popped up into our case directory (bfs_200k_DDES) as soon as the simulation started.
Depending on how far along you are in the simulation, the log.pimplefoam file might be quite long. To give you a quick run through of how it looks, let us visualize the beginning of it:
See log.pimpleFoam
[user@gra-login1] vim log.pimpleFoamStarting time loop
Courant Number mean: 0.37558691 max: 5.0866216Time = 0.0081
PIMPLE: iteration 1DILUPBiCGStab: Solving for Ux, Initial residual = 0.0543257, Final residual = 0.0030492453, No Iterations 1DILUPBiCGStab: Solving for Uy, Initial residual = 0.019132138, Final residual = 0.0009267347, No Iterations 1DILUPBiCGStab: Solving for Uz, Initial residual = 0.066712166, Final residual = 0.0049700607, No Iterations 1GAMG: Solving for p, Initial residual = 0.98050352, Final residual = 0.021488268, No Iterations 2time step continuity errors : sum local = 0.00044866542, global = -1.3391353e-05, cumulative = -1.3391353e-05GAMG: Solving for p, Initial residual = 0.14858043, Final residual = 8.6866034e-07, No Iterations 45time step continuity errors : sum local = 3.0582936e-08, global = -3.2758568e-09, cumulative = -1.3394628e-05PIMPLE: iteration 2DILUPBiCGStab: Solving for Ux, Initial residual = 0.058760738, Final residual = 0.003117804, No Iterations 1DILUPBiCGStab: Solving for Uy, Initial residual = 0.021669178, Final residual = 0.00078736235, No Iterations 1DILUPBiCGStab: Solving for Uz, Initial residual = 0.12996742, Final residual = 0.010186982, No Iterations 1GAMG: Solving for p, Initial residual = 0.59370611, Final residual = 0.014856221, No Iterations 2time step continuity errors : sum local = 0.00034748909, global = 1.1426898e-05, cumulative = -1.9677303e-06GAMG: Solving for p, Initial residual = 0.30867957, Final residual = 9.1670175e-07, No Iterations 49time step continuity errors : sum local = 1.3500471e-08, global = 1.43852e-09, cumulative = -1.9662918e-06PIMPLE: iteration 3DILUPBiCGStab: Solving for Ux, Initial residual = 0.015980338, Final residual = 0.0006615279, No Iterations 1DILUPBiCGStab: Solving for Uy, Initial residual = 0.0087843757, Final residual = 0.00022240436, No Iterations 1DILUPBiCGStab: Solving for Uz, Initial residual = 0.11693161, Final residual = 0.006951935, No Iterations 1GAMG: Solving for p, Initial residual = 0.65043891, Final residual = 0.010075974, No Iterations 2time step continuity errors : sum local = 0.00010854745, global = 2.6004423e-06, cumulative = 6.3415051e-07GAMG: Solving for p, Initial residual = 0.40140073, Final residual = 9.079401e-07, No Iterations 47time step continuity errors : sum local = 4.8308146e-09, global = 5.1590399e-10, cumulative = 6.3466642e-07Important information to retain from the log file are:
- The time iteration corresponds to the time integration of the equations of motion mentioned in section 2.3.
- The CFL or Courant number is displayed at every time stamp.
- Residuals are shown at each iteration for all velocity components and pressure.
- Local and global mass conservation is also printed at each iteration.
These 4 pieces of information are already incredibly useful to understand if the simulation is converging, diverging, if mass is globally conserved, or if there is a problem in the domain.
The simulation will probably run for several hours, and the ideal scenario is that every once in a while we check the behavior of the residuals and mass conservation. As you might guess, staring at numbers on the screen is not the best approach, and, once again, it is better to adopt an automated mechanism to visualize residuals. This can be done using gnuplot, a command-line and GUI program that can generate two- and three-dimensional plots of functions, data, and data fits. Gnuplot is usually present by default in any UNIX system, however, to make sure you have it in your profile on the cluster, you can type:
[user@gra-login1] gnuplot
G N U P L O T Version 5.4 patchlevel 2 last modified 2021-06-01
Copyright (C) 1986-1993, 1998, 2004, 2007-2021 Thomas Williams, Colin Kelley and many others
gnuplot home: http://www.gnuplot.info faq, bugs, etc: type "help FAQ" immediate help: type "help" (plot window: hit 'h')If you do not see the gnuplot welcome message, or if the terminal throws you an error, you can load the gnuplot module just like any other module:
[user@gra-login1] module load gnuplot[user@gra-login1] module saveWe can now write a simple script to plot residuals during runtime:
set logscale yset title "Residuals" font "arial,22"set ylabel "Residual" font "arial,18"set xlabel "Iteration" font "arial,18"set key outsideset grid
nCorrectors=1nNonOrthogonalCorrectors=3
nCont = nCorrectorsnP = (nNonOrthogonalCorrectors+1)*nContnPSkip = nP-1nContSkip = nCont-1
plot "< cat log.pimpleFoam | grep 'Solving for Ux' | cut -d' ' -f9 | tr -d ','" title 'Ux' with linespoints,\"< cat log.pimpleFoam | grep 'Solving for Uy' | cut -d' ' -f9 | tr -d ','" title 'Uy' with linespoints,\"< cat log.pimpleFoam | grep 'Solving for Uz' | cut -d' ' -f9 | tr -d ','" title 'Uz' with linespoints lc 8pause 1xmax = GPVAL_DATA_X_MAX+2xmin = xmax-40set xrange [xmin:xmax]rereadThe script above will plot the residuals for all velocity components during the past 40 time steps. The students can modify the highlighted line in the script to change the plotting range. The script MUST BE located in the same directory of the log.pimpleFoam, and to run it simply type:
[user@gra-login1] gnuplot plot-residuals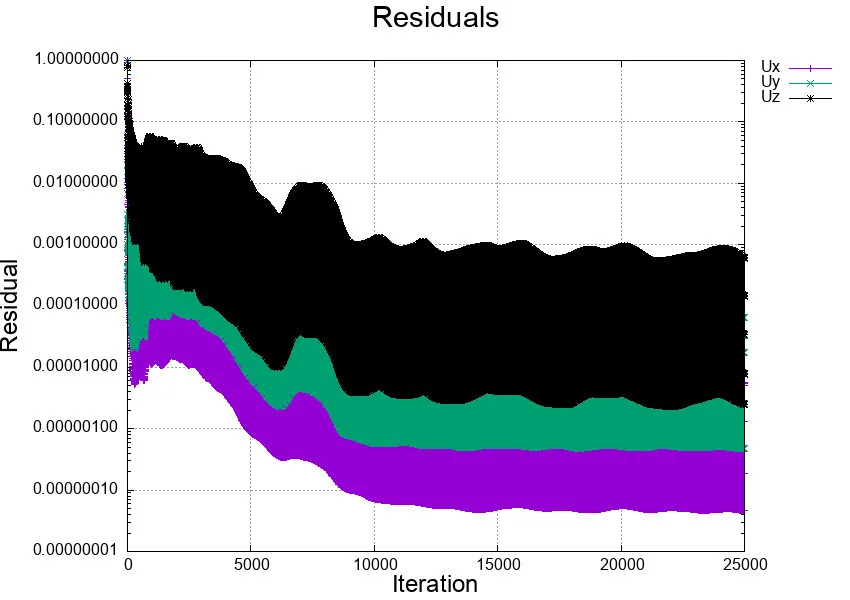
If you recall, in lines 252 to 255 of Backstep_str_config.cfg file, we have instructed SU2 to generate an output file containing convergence history under the name, history.csv. If you notice, this file popped up into our case directory (02_BFS_SU2/Coarse/) as soon as the simulation started.
Depending on how far along in the simulation you are, the history.csv file might be quite long. To give you a quick run through of how it looks, let us visualize the beginning of it:
See history.csv
1 "Time_Iter","Outer_Iter","Inner_Iter", "rms[P]" , "rms[U]" , "rms[V]" , "rms[W]" , "rms[nu]"2 0, 0, 4, -8.36488691, -8.323185486, -8.749827447, -6.595921079, -13.984444693 1, 0, 4, -8.12803185, -8.128927621, -8.208809081, -6.192896793, -14.065655284 2, 0, 4, -7.960896066, -7.959261575, -7.869191588, -5.866127477, -14.204453585 3, 0, 4, -7.87770485, -7.865723278, -7.730622795, -5.674950425, -14.332125746 4, 0, 4, -7.851101629, -7.795018166, -7.707723687, -5.578550417, -14.394996837 5, 0, 4, -7.848972586, -7.697854479, -7.734610289, -5.498682789, -14.512603348 6, 0, 4, -7.839963907, -7.596020898, -7.758581592, -5.383722204, -14.467932389 7, 0, 4, -7.84740461, -7.54672322, -7.79919976, -5.3481669, -14.4535115310 8, 0, 4, -7.903200003, -7.571866226, -7.89494942, -5.392784656, -14.54477387In this case, the file contains the root-mean-square
-
Screen output: The convergence history printed on the console.
-
History output: The convergence history written to a file.
-
Volume output: Everything was written to the visualization and restart files.
-
Output field: A single scalar value for screen and history output or a vector of a scalar quantity at every node in the mesh for volume output.
-
Output group: A collection of output fields.
More information can be found HERE. The simulation will be running probably for several hours, and the ideal scenario is that every once in a while we check the behavior of the
[user@gra-login1] gnuplot
G N U P L O T Version 5.4 patchlevel 2 last modified 2021-06-01
Copyright (C) 1986-1993, 1998, 2004, 2007-2021 Thomas Williams, Colin Kelley and many others
gnuplot home: http://www.gnuplot.info faq, bugs, etc: type "help FAQ" immediate help: type "help" (plot window: hit 'h')If you do not see the gnuplot welcome message, or if the terminal throws you an error, you can load the gnuplot module just like any other module:
[user@gra-login1] module load gnuplotWe can now write a simple script to plot
set datafile separator ","set title "Residuals" font "arial,22"set ylabel "log10 of Residual" font "arial,18"set xlabel "Iteration" font "arial,18"set key autotitle columnheadset grid
plot "history.csv" using 1:5 with lines, "history.csv" using 1:6 with lines, "history.csv" using 1:7 with linespause 1if (GPVAL_DATA_X_MAX<=500) { xmax = 550}else { xmax = GPVAL_DATA_X_MAX+50}xmin = 0ymax = GPVAL_DATA_Y_MAX+1ymin = GPVAL_DATA_Y_MIN-1set xrange [xmin:xmax]set yrange [ymin:ymax]rereadThe students can modify the highlighted lines in the script above to change the plotting range. The script MUST BE located in the same directory of the lhistory.csv, and to run it simply type:
[user@gra-login1] gnuplot plot-residuals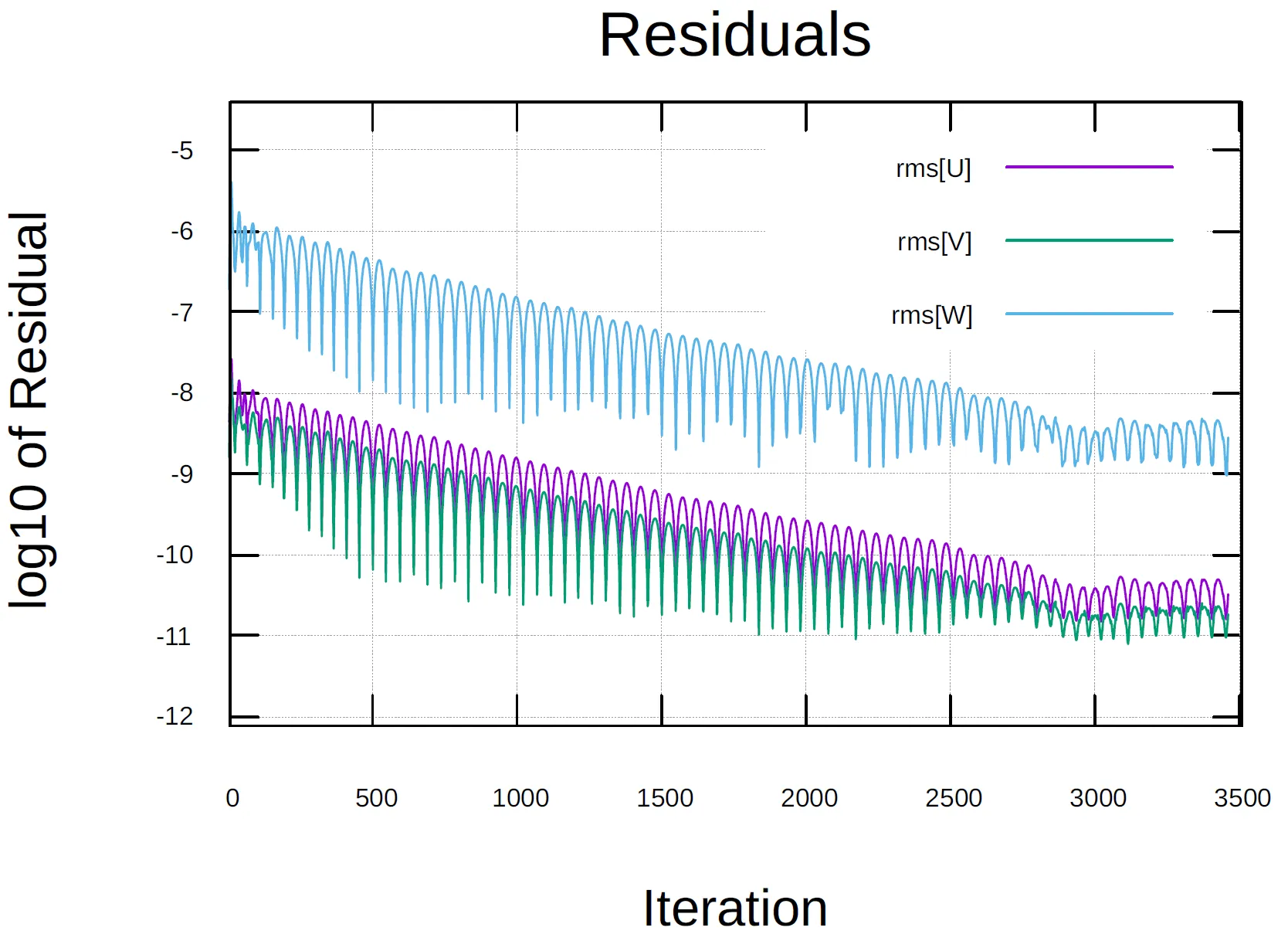
You might want to think about output files
When running a simulation in parallel, it is crucial to think about the impact of the output files on the HPC workflow. Running
Number of output files
Depending on the CFD tool used, when running a simulation in parallel, we need to remember that the computational domain has been decomposed into /scratch directory.
In simple terms, the coarse simulation of the BFS we have just carried out on 64 processors for about 25000 iterations would generate about 8 million files!! This is why the time interval between snapshots should be chosen wisely.
This type of output is known as parallel output, and one should always consider merging all processors’ files after the simulation is done or (if possible) during runtime. This is precisely why reconstructPar was included in the OpenFOAM batch script file.
Although some available CFD tools will perform this operation by default, it is always a good idea to perform a rough estimate of the number of output files expected from a numerical simulation. Let’s consider our BFS simulation over a total time of
- Number of files parallel output:
- Number of files merged output:
Check the documentation of your CFD tool as you might be able to change the way output files are written. In OpenFOAM, for instance, you can switch between the two write methods on the fly by modifying the controlDict entry while your case is running.
Size of output files
Some thought should also be given to the size of the output files. Without going int0 too much detail, in HPC we have two possible output formats:
-
Binary: as mentioned in section 1 of this course, the binary language is very efficient for programs and is not designed for humans to read. Executables for instance are written in binary code by the compiler, and contain the set of instructions a program has to execute.
-
ASCII: stands for American Standard Code for Information Interchange. It is a coded character set consisting of 128 7-bit characters. There are 32 control characters, 94 graphic characters, the space character, and the delete character. ASCII file format that can be easily read by humans. A very common text file (.txt) is an ASCII file.
Why does this matter in CFD and HPC?
Binary format is faster for read/write since the machine does not have to convert to a human-readable format. The size of a binary output file is also smaller as compared to an ASCII file. Most binary formats are platform-dependent and not easily transferable between systems.
Applying this reasoning to a CFD case:
-
For complex geometries and very large mesh files where the goal is to print the output for hundreds or thousands of snapshots, binary would be a better choice, as writing many data points and many snapshots can be done relatively instantaneously (compared to converting and writing thousands of ASCII files).
-
For relatively small cases on simple geometries, where the number of output files is not too large, writing ASCII files will not cause a significant performance hit, and one can open and manipulate single output files.
- ASCII:
- Suitable for small meshes and few snapshots.
- Can be visualized and edited using regular text editors.
- Binary:
- Smaller file size.
- Faster to read/write
- Suitable for large meshes and complex geometries.
- ASCII:
- Larger file size.
- Binary:
- Cannot be read and edited by regular text editors.
Align restarts with clock time
Many HPC systems use fair share schedulers which often impose maximum time durations for each submitted job. On most Digital Research Alliance clusters, the maximum walltime of a job is 24 hours. Therefore, restart files (also called breakpoint) need to be output in order to run the simulation for more than 24 hours. To optimize the computational usage, we should seek to have a restart file written immediately before the end of the simulation.
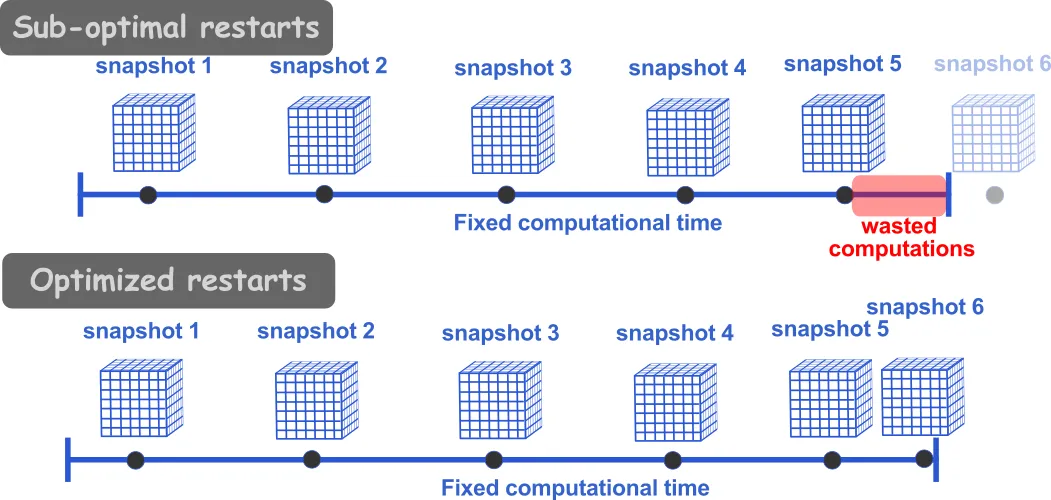
For a 24 h run (86,400 seconds), if we know that the writing files take about 2 minutes (dependent on the size of the simulation), then we would want to have a restart file written at, say 86,000 seconds of wallclock time. Additionally, if we want to assure that we have some intermediary restart files (in case of issues with the simulation), we may want to write every 8,600 seconds of wallclock time. In openFoam this can be done by setting the following in the controlDict file:
writeControl clockTime;writeInterval 8600;Can I trust my results?
Before diving too much into visualization sessions of our fresh CFD data, we should always ask ourselves if we can trust our results? This is a crucial question to answer as it is central to answering our scientific question(s). In order trust our CFD results, there are two important aspects to consider:
Grid sensitivity study
This is an internal test that provides an assessment that the results are insensitive to selected computational grid. For highly non-linear problems, strongly influenced by the boundary conditions, the grid sensitivity study is carried out by running 3 or more simulations of the same problem, with the same boundary and initial conditions, on successively refined grids, ideally by doubling the refinement in each direction. If the numerical method is stable, and all approximations used in the discretization (finite differences, finite volumes, etc.) are consistent, you will find that the solution eventually converges to a grid-insensitive solution (Ferziger and Peric, 2002). Only at this point can we conclude that the numerical method and discretization scheme chosen can be trusted.
As an example, below we show the result of a grid sensitivity study on the BFS example we used in this course. Although we only showed how to run the coarsest mesh, two finer grids (400k, and 800k) have been tested. The figure shows the time-averaged streamwise velocity
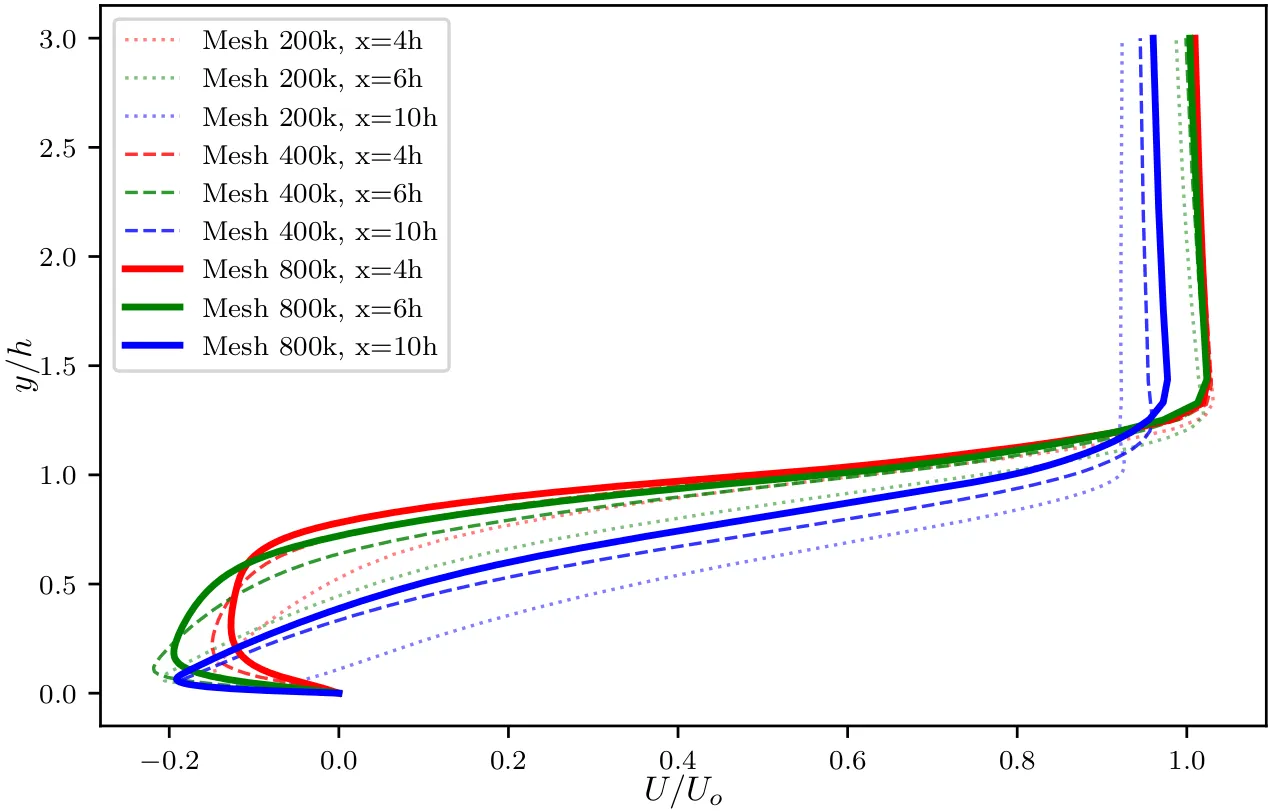
In the figure above, we notice that the dash and solid curves at all locations (different colors) are very close to each other. One can estimate the error between the two profiles at each location, and if the difference between the profiles is less than 5% we can conclude that grid convergence has been reached on the medium-fine grid of about 400,000 points. Any simulation on a grid finer than this will not constitute a significant improvement on the solution and will therefore be an ineffective use of computational resources.
Verification and validation check
This is an external test where the goal is to assess the the models are correctly implemented (verification) and the accuracy of the models to represent real-world flow (validation). The verification and validation processes are well established and standardized among the CFD community. We define:
- Verification is “The process of determining that a model implementation accurately represents the developer’s conceptual description of the model and the solution to the model. (AIAA G-077-1998)“. The objective is to assess that the implementation of the conceptual model in the solver is correct.
- Validation is “The process of determining the degree to which a model is an accurate representation of the real world from the perspective of the intended uses of the model. (AIAA G-077-1998)“. The objective is to assure that the selected model represents the physical reality.
For well-established codes, verification of the implemented models is usually done prior to the codes release (see e.g. openFoam’s V&V). Validation, on the other hand, is a critical step to assure the validity and gain confidence in the CFD results. As CFD is predictive tool, it is often difficult to validate the specific test case. In those cases, there are many canonical flows whose sole purpose is to serve as benchmark (lead driven cavity flow, backward facing step, turbulent boundary layer, periodic channel flow, etc.).
Once you assure that the CFD solution is grid-independent, the CFD tool and models are verified, and the CFD tools validated, then we can gain trust in the CFD results and can advance the understanding of a physical phenomenon.
Let us visualize the flow!
(more on this in the next class)
Having finished this class, you should now be able to answer the following questions:
- How do I organize simulation files on the cluster?
- How do I run a large-scale CFD simulation on HPC systems?
- How do I monitor the simulation during runtime?
- How do I save data efficiently?